Kaist
Korean

Students News
-

Machine Learning-Based Algorithm to Speed up DNA S..
The algorithm presents the first full-fledged, short-read alignment software that leverages learned indices for solving the exact match search problem for efficient seeding < Image:Scientists from KAIST develops new machine-learning-based approach to speed up DNA sequencing. > The human genome consists of a complete set of DNA, which is about 6.4 billion letters long. Because of its size, reading the whole genome sequence at once is challenging. So scientists use DNA sequencers to produce hundreds of millions of DNA sequence fragments, or short reads, up to 300 letters long. Then the DNA sequencer assembles all the short reads like a giant jigsaw puzzle to reconstruct the entire genome sequence. Even with very fast computers, this job can take hours to complete. A research team at KAIST has achieved up to 3.45x faster speeds by developing the first short-read alignment software that uses a recent advance in machine-learning called a learned index. The research team reported their findings on March 7, 2022 in the journal Bioinformatics. The software has been released as open source and can be found on github (https://github.com/kaist-ina/BWA-MEME). Next-generation sequencing (NGS) is a state-of-the-art DNA sequencing method. Projects are underway with the goal of producing genome sequencing at population scale. Modern NGS hardware is capable of generating billions of short reads in a single run. Then the short reads have to be aligned with the reference DNA sequence. With large-scale DNA sequencing operations running hundreds of next-generation sequences, the need for an efficient short read alignment tool has become even more critical. Accelerating the DNA sequence alignment would be a step toward achieving the goal of population-scale sequencing. However, existing algorithms are limited in their performance because of their frequent memory accesses. BWA-MEM2 is a popular short-read alignment software package currently used to sequence the DNA. However, it has its limitations. The state-of-the-art alignment has two phases – seeding and extending. During the seeding phase, searches find exact matches of short reads in the reference DNA sequence. During the extending phase, the short reads from the seeding phase are extended. In the current process, bottlenecks occur in the seeding phase. Finding the exact matches slows the process. The researchers set out to solve the problem of accelerating the DNA sequence alignment. To speed the process, they applied machine learning techniques to create an algorithmic improvement. Their algorithm, BWA-MEME (BWA-MEM emulated) leverages learned indices to solve the exact match search problem. The original software compared one character at a time for an exact match search. The team’s new algorithm achieves up to 3.45x faster speeds in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60x and memory accesses by 8.77x. “Through this study, it has been shown that full genome big data analysis can be performed faster and less costly than conventional methods by applying machine learning technology,” said Professor Dongsu Han from the School of Electrical Engineering at KAIST. The researchers’ ultimate goal was to develop efficient software that scientists from academia and industry could use on a daily basis for analyzing big data in genomics. “With the recent advances in artificial intelligence and machine learning, we see so many opportunities for designing better software for genomic data analysis. The potential is there for accelerating existing analysis as well as enabling new types of analysis, and our goal is to develop such software,” added Han. Whole genome sequencing has traditionally been used for discovering genomic mutations and identifying the root causes of diseases, which leads to the discovery and development of new drugs and cures. There could be many potential applications. Whole genome sequencing is used not only for research, but also for clinical purposes. “The science and technology for analyzing genomic data is making rapid progress to make it more accessible for scientists and patients. This will enhance our understanding about diseases and develop a better cure for patients of various diseases.” The research was funded by the National Research Foundation of the Korean government’s Ministry of Science and ICT. -Publication Youngmok Jung, Dongsu Han, “BWA-MEME:BWA-MEM emulated with a machine learning approach,” Bioinformatics, Volume 38, Issue 9, May 2022 (https://doi.org/10.1093/bioinformatics/btac137) -Profile Professor Dongsu Han School of Electrical Engineering KAIST
-

Professor June-Koo Rhee’s Team Wins the QHack Open..
< From left: Ju-young Ryu, Jeung-rak Lee, and Eyuel Elala in Professor June-Koo Rhee > The research team consisting of three master students Ju-Young Ryu, Jeung-rak Lee, and Eyel Elala in Professor June-Koo Rhee’s group from the KAIST IRTC of Quantum Computing for AI has won the first place at the QHack 2022 Open Hackathon Science Challenge. The QHack 2022 Open Hackathon is one of the world’s prestigious quantum software hackathon events held by US Xanadu, in which 250 people from 100 countries participate. Major sponsors such as IBM Quantum, AWS, CERN QTI, and Google Quantum AI proposed challenging problems, and a winning team is selected judged on team projects in each of the 13 challenges. The KAIST team supervised by Professor Rhee received the First Place prize on the Science Challenge which was organized by the CERN QTI of the European Communities. The team will be awarded an opportunity to tour CERN’s research lab in Europe for one week along with an online internship. The students on the team presented a method for “Leaning Based Error Mitigation for VQE,” in which they implemented an LBEM protocol to lower the error in quantum computing, and leveraged the protocol in the VQU algorithm which is used to calculate the ground state energy of a given molecule. Their research successfully demonstrated the ability to effectively mitigate the error in IBM Quantum hardware and the virtual error model. In conjunction, Professor June-Koo (Kevin) Rhee founded a quantum computing venture start-up, Qunova Computing(https://qunovacomputing.com), with technology tranfer from the KAIST ITRC of Quantum Computing for AI. Qunova Computing is one of the frontier of the quantum software industry in Korea.
-

Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain < Figure:Experimental paradigm. Subjects were instructed to perform reach-and-grasp movements to designate the locations of the target in three-dimensional space. (a) Subjects A and B were provided the visual cue as a real tennis ball at one of four pseudo-randomized locations. (b) Subjects A and B were provided the visual cue as a virtual reality clip showing a sequence of five stages of a reach-and-grasp movement. > Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI). A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a robotic limb. There are two main techniques for monitoring neural signals in BMIs: electroencephalography (EEG) and electrocorticography (ECoG). The EEG exhibits signals from electrodes on the surface of the scalp and is widely employed because it is non-invasive, relatively cheap, safe and easy to use. However, the EEG has low spatial resolution and detects irrelevant neural signals, which makes it difficult to interpret the intentions of individuals from the EEG. On the other hand, the ECoG is an invasive method that involves placing electrodes directly on the surface of the cerebral cortex below the scalp. Compared with the EEG, the ECoG can monitor neural signals with much higher spatial resolution and less background noise. However, this technique has several drawbacks. “The ECoG is primarily used to find potential sources of epileptic seizures, meaning the electrodes are placed in different locations for different patients and may not be in the optimal regions of the brain for detecting sensory and movement signals,” explained Professor Jaeseung Jeong, a brain scientist at KAIST. “This inconsistency makes it difficult to decode brain signals to predict movements.” To overcome these problems, Professor Jeong’s team developed a new method for decoding ECoG neural signals during arm movement. The system is based on a machine-learning system for analysing and predicting neural signals called an ‘echo-state network’ and a mathematical probability model called the Gaussian distribution. In the study, the researchers recorded ECoG signals from four individuals with epilepsy while they were performing a reach-and-grasp task. Because the ECoG electrodes were placed according to the potential sources of each patient’s epileptic seizures, only 22% to 44% of the electrodes were located in the regions of the brain responsible for controlling movement. During the movement task, the participants were given visual cues, either by placing a real tennis ball in front of them, or via a virtual reality headset showing a clip of a human arm reaching forward in first-person view. They were asked to reach forward, grasp an object, then return their hand and release the object, while wearing motion sensors on their wrists and fingers. In a second task, they were instructed to imagine reaching forward without moving their arms. The researchers monitored the signals from the ECoG electrodes during real and imaginary arm movements, and tested whether the new system could predict the direction of this movement from the neural signals. They found that the novel decoder successfully classified arm movements in 24 directions in three-dimensional space, both in the real and virtual tasks, and that the results were at least five times more accurate than chance. They also used a computer simulation to show that the novel ECoG decoder could control the movements of a robotic arm. Overall, the results suggest that the new machine learning-based BCI system successfully used ECoG signals to interpret the direction of the intended movements. The next steps will be to improve the accuracy and efficiency of the decoder. In the future, it could be used in a real-time BMI device to help people with movement or sensory impairments. This research was supported by the KAIST Global Singularity Research Program of 2021, Brain Research Program of the National Research Foundation of Korea funded by the Ministry of Science, ICT, and Future Planning, and the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education. -Publication Hoon-Hee Kim, Jaeseung Jeong, “An electrocorticographic decoder for arm movement for brain-machine interface using an echo state network and Gaussian readout,” Applied Soft Computing online December 31, 2021 (doi.org/10.1016/j.asoc.2021.108393) -Profile Professor Jaeseung Jeong Department of Bio and Brain Engineering College of Engineering KAIST
-
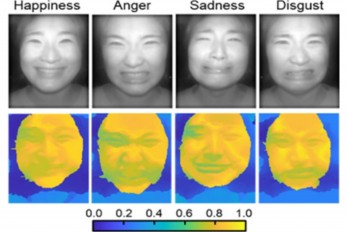
AI Light-Field Camera Reads 3D Facial Expressions
Machine-learned, light-field camera reads facial expressions from high-contrast illumination invariant 3D facial images < Image: Facial expression reading based on MLP classification from 3D depth maps and 2D images obtained by NIR-LFC > A joint research team led by Professors Ki-Hun Jeong and Doheon Lee from the KAIST Department of Bio and Brain Engineering reported the development of a technique for facial expression detection by merging near-infrared light-field camera techniques with artificial intelligence (AI) technology. Unlike a conventional camera, the light-field camera contains micro-lens arrays in front of the image sensor, which makes the camera small enough to fit into a smart phone, while allowing it to acquire the spatial and directional information of the light with a single shot. The technique has received attention as it can reconstruct images in a variety of ways including multi-views, refocusing, and 3D image acquisition, giving rise to many potential applications. However, the optical crosstalk between shadows caused by external light sources in the environment and the micro-lens has limited existing light-field cameras from being able to provide accurate image contrast and 3D reconstruction. The joint research team applied a vertical-cavity surface-emitting laser (VCSEL) in the near-IR range to stabilize the accuracy of 3D image reconstruction that previously depended on environmental light. When an external light source is shone on a face at 0-, 30-, and 60-degree angles, the light field camera reduces 54% of image reconstruction errors. Additionally, by inserting a light-absorbing layer for visible and near-IR wavelengths between the micro-lens arrays, the team could minimize optical crosstalk while increasing the image contrast by 2.1 times. Through this technique, the team could overcome the limitations of existing light-field cameras and was able to develop their NIR-based light-field camera (NIR-LFC), optimized for the 3D image reconstruction of facial expressions. Using the NIR-LFC, the team acquired high-quality 3D reconstruction images of facial expressions expressing various emotions regardless of the lighting conditions of the surrounding environment. The facial expressions in the acquired 3D images were distinguished through machine learning with an average of 85% accuracy – a statistically significant figure compared to when 2D images were used. Furthermore, by calculating the interdependency of distance information that varies with facial expression in 3D images, the team could identify the information a light-field camera utilizes to distinguish human expressions. Professor Ki-Hun Jeong said, “The sub-miniature light-field camera developed by the research team has the potential to become the new platform to quantitatively analyze the facial expressions and emotions of humans.” To highlight the significance of this research, he added, “It could be applied in various fields including mobile healthcare, field diagnosis, social cognition, and human-machine interactions.” This research was published in Advanced Intelligent Systems online on December 16, under the title, “Machine-Learned Light-field Camera that Reads Facial Expression from High-Contrast and Illumination Invariant 3D Facial Images.” This research was funded by the Ministry of Science and ICT and the Ministry of Trade, Industry and Energy. -Publication “Machine-learned light-field camera that reads fascial expression from high-contrast and illumination invariant 3D facial images,” Sang-In Bae, Sangyeon Lee, Jae-Myeong Kwon, Hyun-Kyung Kim. Kyung-Won Jang, Doheon Lee, Ki-Hun Jeong, Advanced Intelligent Systems, December 16, 2021 (doi.org/10.1002/aisy.202100182) -Profile Professor Ki-Hun Jeong Biophotonic Laboratory Department of Bio and Brain Engineering KAIST Professor Doheon Lee Department of Bio and Brain Engineering KAIST
-

Face Detection in Untrained Deep Neural Networks
A KAIST team shows that primitive visual selectivity of faces can arise spontaneously in completely untrained deep neural networks Researchers have found that higher visual cognitive functions can arise spontaneously in untrained neural networks. A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has shown that visual selectivity of facial images can arise even in completely untrained deep neural networks. This new finding has provided revelatory insights into mechanisms underlying the development of cognitive functions in both biological and artificial neural networks, also making a significant impact on our understanding of the origin of early brain functions before sensory experiences. The study published in Nature Communications on December 16 demonstrates that neuronal activities selective to facial images are observed in randomly initialized deep neural networks in the complete absence of learning, and that they show the characteristics of those observed in biological brains. The ability to identify and recognize faces is a crucial function for social behavior, and this ability is thought to originate from neuronal tuning at the single or multi-neuronal level. Neurons that selectively respond to faces are observed in young animals of various species, and this raises intense debate whether face-selective neurons can arise innately in the brain or if they require visual experience. Using a model neural network that captures properties of the ventral stream of the visual cortex, the research team found that face-selectivity can emerge spontaneously from random feedforward wirings in untrained deep neural networks. The team showed that the character of this innate face-selectivity is comparable to that observed with face-selective neurons in the brain, and that this spontaneous neuronal tuning for faces enables the network to perform face detection tasks. These results imply a possible scenario in which the random feedforward connections that develop in early, untrained networks may be sufficient for initializing primitive visual cognitive functions. Professor Paik said, “Our findings suggest that innate cognitive functions can emerge spontaneously from the statistical complexity embedded in the hierarchical feedforward projection circuitry, even in the complete absence of learning”. He continued, “Our results provide a broad conceptual advance as well as advanced insight into the mechanisms underlying the development of innate functions in both biological and artificial neural networks, which may unravel the mystery of the generation and evolution of intelligence.” This work was supported by the National Research Foundation of Korea (NRF) and by the KAIST singularity research project. -Publication Seungdae Baek, Min Song, Jaeson Jang, Gwangsu Kim, and Se-Bum Baik, “Face detection in untrained deep neural network,” Nature Communications 12, 7328 on Dec.16, 2021 (https://doi.org/10.1038/s41467-021-27606-9) -Profile Professor Se-Bum Paik Visual System and Neural Network Laboratory Program of Brain and Cognitive Engineering Department of Bio and Brain Engineering College of Engineering KAIST
-

Team KAIST to Race at CES 2022 Autonomous Challeng..
Five top university autonomous racing teams will compete in a head-to-head passing competition in Las Vegas < Team KAIST of self-driving team from the Unmanned System Research Group (USRG) advised by Professor Hyunchul Shim (second from right) > A self-driving racing team from the KAIST Unmanned System Research Group (USRG) advised by Professor Hyunchul Shim will compete at the Autonomous Challenge at the Consumer Electronic Show (CES) on January 7, 2022. The head-to-head, high speed autonomous racecar passing competition at the Las Vegas Motor Speedway will feature the finalists and semifinalists from the Indy Autonomous Challenge in October of this year. Team KAIST qualified as a semifinalist at the Indy Autonomous Challenge and will join four other university teams including the winner of the competition, Technische Universität München. Team KAIST’s AV-21 vehicle is capable of driving on its own at more than 200km/h will be expected to show a speed of more than 300 km/h at the race.The participating teams are: 1. KAIST 2. EuroRacing : University of Modena and Reggio Emilia (Italy), University of Pisa (Italy), ETH Zürich (Switzerland), Polish Academy of Sciences (Poland) 3. MIT-PITT-RW, Massachusetts Institute of Technology, University of Pittsburgh, Rochester Institute of Technology, University of Waterloo (Canada) 4.PoliMOVE – Politecnico di Milano (Italy), University of Alabama 5.TUM Autonomous Motorsport – Technische Universität München (Germany) Professor Shim’s team is dedicated to the development and validation of cutting edge technologies for highly autonomous vehicles. In recognition of his pioneering research in unmanned system technologies, Professor Shim was honored with the Grand Prize of the Minister of Science and ICT on December 9. “We began autonomous vehicle research in 2009 when we signed up for Hyundai Motor Company’s Autonomous Driving Challenge. For this, we developed a complete set of in-house technologies such as low-level vehicle control, perception, localization, and decision making.” In 2019, the team came in third place in the Challenge and they finally won this year. For years, his team has participated in many unmanned systems challenges at home and abroad, gaining recognition around the world. The team won the inaugural 2016 IROS autonomous drone racing and placed second in the 2018 IROS Autonomous Drone Racing Competition. They also competed in 2017 MBZIRC, ranking fourth in Missions 2 and 3, and fifth in the Grand Challenge. Most recently, the team won the first round of Lockheed Martin’s Alpha Pilot AI Drone Innovation Challenge. The team is now participating in the DARPA Subterranean Challenge as a member of Team CoSTAR with NASA JPL, MIT, and Caltech. “We have accumulated plenty of first-hand experience developing autonomous vehicles with the support of domestic companies such as Hyundai Motor Company, Samsung, LG, and NAVER. In 2017, the autonomous vehicle platform “EureCar” that we developed in-house was authorized by the Korean government to lawfully conduct autonomous driving experiment on public roads,” said Professor Shim. The team has developed various key technologies and algorithms related to unmanned systems that can be categorized into three major components: perception, planning, and control. Considering the characteristics of the algorithms that make up each module, their technology operates using a distributed computing system. Since 2015, the team has been actively using deep learning algorithms in the form of perception subsystems. Contextual information extracted from multi-modal sensory data gathered via cameras, lidar, radar, GPS, IMU, etc. is forwarded to the planning subsystem. The planning module is responsible for the decision making and planning required for autonomous driving such as lane change determination and trajectory planning, emergency stops, and velocity command generation. The results from the planner are fed into the controller to follow the planned high-level command. The team has also developed and verified the possibility of an end-to-end deep learning based autonomous driving approach that replaces a complex system with one single AI network.
-
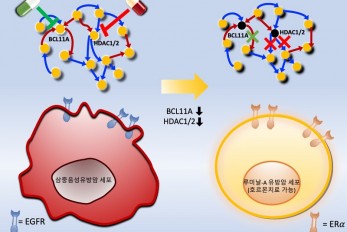
Connecting the Dots to Find New Treatments for Bre..
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers < Professor Kwang-Hyun Cho and colleagues have developed a mathematical model and identified optimal targets reprogramming basal-like cancer cells into hormone therapy-responsive luminal-A cells by deciphering the complex molecular interactions within these cells through a systems biological approach. > Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research. Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα). KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment. To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off. “There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.” Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans. “Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho. The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones. This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project. -Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine Therapy Vulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621) -Profile Professor Kwang-Hyun Cho Laboratory for Systems Biology and Bio-Inspired Engineering Department of Bio and Brain Engineering KAIST
-

A Team of Three PhD Candidates Wins the Korea Semi..
“We felt a sense of responsibility to help the nation advance its semiconductor design technology” < PhD candidates at the School of Electrical Engineering Yoon-Seo Cho, Sun-Eui Park, and Ju-Eun Bang (from left) > A CMOS (complementary metal-oxide semiconductor)-based “ultra-low noise signal chip” for 6G communications designed by three PhD candidates at the KAIST School of Electrical Engineering won the Presidential Award at the 22nd Korea Semiconductor Design Contest. The winners are PhD candidates Sun-Eui Park, Yoon-Seo Cho, and Ju-Eun Bang from the Integrated Circuits and System Lab run by Professor Jaehyouk Choi. The contest, which is hosted by the Ministry of Trade, Industry and Energy and the Korea Semiconductors Industry Association, is one of the top national semiconductor design contests for college students. Park said the team felt a sense of responsibility to help advance semiconductor design technology in Korea when deciding to participate the contest. The team expressed deep gratitude to Professor Choi for guiding their research on 6G communications. “Our colleagues from other labs and seniors who already graduated helped us a great deal, so we owe them a lot,” explained Park. Cho added that their hard work finally got recognized and that acknowledgement pushes her to move forward with her research. Meanwhile, Bang said she is delighted to see that many people seem to be interested in her research topic. Research for 6G is attempting to reach 1 tera bps (Tbps), 50 times faster than 5G communications with transmission speeds of up to 20 gigabytes. In general, the wider the communication frequency band, the higher the data transmission speed. Thus, the use of frequency bands above 100 gigahertz is essential for delivering high data transmission speeds for 6G communications. However, it remains a big challenge to make a precise benchmark signal that can be used as a carrier wave in a high frequency band. Despite the advantages of CMOS’s ultra-small and low-power design, it still has limitations at high frequency bands and its operating frequency. Thus, it was difficult to achieve a frequency band above 100 gigahertz. To overcome these challenges, the three students introduced ultra-low noise signal generation technology that can support high-order modulation technologies. This technology is expected to contribute to increasing the price competitiveness and density of 6G communication chips that will be used in the future. 5G just got started in 2020 and still has long way to go for full commercialization. Nevertheless, many researchers have started preparing for 6G technology, targeting 2030 since a new cellular communication appears in every other decade. Professor Choi said, “Generating ultra-high frequency signals in bands above 100 GHz with highly accurate timing is one of the key technologies for implementing 6G communication hardware. Our research is significant for the development of the world’s first semiconductor chip that will use the CMOS process to achieve noise performance of less than 80fs in a frequency band above 100 GHz.” The team members plan to work as circuit designers in Korean semiconductor companies after graduation. “We will continue to research the development of signal generators on the topic of award-winning 6G. We would like to continue our research on high-speed circuit designs such as ultra-fast analog-to-digital converters,” Park added.
-

Professor Sung-Ju Lee’s Team Wins the Best Paper a..
< Professor Sung-Ju Lee, Professor Eun Kyoung, Choe Hyunsung Cho, Daeun Choi, Donghwi Kim, Wan Ju Kang (from left) > A research team led by Professor Sung-Ju Lee at the School of Electrical Engineering won the Best Paper Award and the Methods Recognition Award from ACM CSCW (International Conference on Computer-Supported Cooperative Work and Social Computing) 2021 for their paper “Reflect, not Regret: Understanding Regretful Smartphone Use with App Feature-Level Analysis”. Founded in 1986, CSCW has been a premier conference on HCI (Human Computer Interaction) and Social Computing. This year, 340 full papers were presented and the best paper awards are given to the top 1% papers of the submitted. Methods Recognition, which is a new award, is given “for strong examples of work that includes well developed, explained, or implemented methods, and methodological innovation.” Hyunsung Cho (KAIST alumus and currently a PhD candidate at Carnegie Mellon University), Daeun Choi (KAIST undergraduate researcher), Donghwi Kim (KAIST PhD Candidate), Wan Ju Kang (KAIST PhD Candidate), and Professor Eun Kyoung Choe (University of Maryland and KAIST alumna) collaborated on this research. The authors developed a tool that tracks and analyzes which features of a mobile app (e.g., Instagram’s following post, following story, recommended post, post upload, direct messaging, etc.) are in use based on a smartphone’s User Interface (UI) layout. Utilizing this novel method, the authors revealed which feature usage patterns result in regretful smartphone use. Professor Lee said, “Although many people enjoy the benefits of smartphones, issues have emerged from the overuse of smartphones. With this feature level analysis, users can reflect on their smartphone usage based on finer grained analysis and this could contribute to digital wellbeing.” < Research achievements diagram : Application feature level usage analysis / UI LAYOUT Analysis >
-

A Study Shows Reactive Electrolyte Additives Impro..
Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives provide an opportunity to improve high-performance lithium metal batteries < A combination of lithium difluoro (bisoxalato) phosphate as an F donor and lithium nitrate as an N donor with different electron accepting abilities and adsorption tendencies improves the cycle performance of Li|NCM811 full cells through the creation of a dual-layer SEI on a Li metal anode and a protective CEI on a Ni-rich cathode. > A research team showed that electrolyte additives increase the lifetime of lithium metal batteries and remarkably improve the performance of fast charging and discharging. Professor Nam-Soon Choi’s team from the Department of Chemical and Biomolecular Engineering at KAIST hierarchized the solid electrolyte interphase to make a dual-layer structure and showed groundbreaking run times for lithium metal batteries. The team applied two electrolyte additives that have different reduction and adsorption properties to improve the functionality of the dual-layer solid electrolyte interphase. In addition, the team has confirmed that the structural stability of the nickel-rich cathode was achieved through the formation of a thin protective layer on the cathode. This study was reported in Energy Storage Materials. Securing high-energy-density lithium metal batteries with a long lifespan and fast charging performance is vital for realizing their ubiquitous use as superior power sources for electric vehicles. Lithium metal batteries comprise a lithium metal anode that delivers 10 times higher capacity than the graphite anodes in lithium-ion batteries. Therefore, lithium metal is an indispensable anode material for realizing high-energy rechargeable batteries. However, undesirable reactions among the electrolytes with lithium metal anodes can reduce the power and this remains an impediment to achieving a longer battery lifespan. Previous studies only focused on the formation of the solid electrolyte interphase on the surface of the lithium metal anode. The team designed a way to create a dual-layer solid electrolyte interphase to resolve the instability of the lithium metal anode by using electrolyte additives, depending on their electron accepting ability and adsorption tendencies. This hierarchical structure of the solid electrolyte interphase on the lithium metal anode has the potential to be further applied to lithium-alloy anodes, lithium storage structures, and anode-free technology to meet market expectations for electrolyte technology. The batteries with lithium metal anodes and nickel-rich cathodes represented 80.9% of the initial capacity after 600 cycles and achieved a high Coulombic efficiency of 99.94%. These remarkable results contributed to the development of protective dual-layer solid electrolyte interphase technology for lithium metal anodes. Professor Choi said that the research suggests a new direction for the development of electrolyte additives to regulate the unstable lithium metal anode-electrolyte interface, the biggest hurdle in research on lithium metal batteries. She added that anode-free secondary battery technology is expected to be a game changer in the secondary battery market and electrolyte additive technology will contribute to the enhancement of anode-free secondary batteries through the stabilization of lithium metal anodes. This research was funded by the Technology Development Program to Solve Climate Change of the National Research Foundation in Korea funded by the Ministry of Science, ICT & Future Planning and the Technology Innovation Program funded by the Ministry of Trade, Industry & Energy, and Hyundai Motor Company. - Publication Saehun Kim, Sung O Park, Min-Young Lee, Jeong-A Lee, Imanuel Kristanto, Tae Kyung Lee, Daeyeon Hwang, Juyoung Kim, Tae-Ung Wi, Hyun-Wook Lee, Sang Kyu Kwak, and Nam Soon Choi, “Stable electrode-electrolyte interfaces constructed by fluorine- and nitrogen-donating ionic additives for high-performance lithium metal batteries,” Energy Storage Materials, 45, 1-13 (2022), (doi: https://doi.org/10.1016/j.ensm.2021.10.031) - Profile Professor Nam-Soon Choi Energy Materials Laboratory Department of Chemical and Biomolecular Engineering KAIST
-

3 PhD Candidates Selected as the 2021 Google Fello..
< The 2021 Google PhD fellow Soo Ye Kim, Sanghyun Woo, and Hae Beom Lee (from left) > PhD candidates Soo Ye Kim and Sanghyun Woo from the KAIST School of Electrical Engineering and Hae Beom Lee from the Kim Jaechul Graduate School of AI were selected as the 2021 Google PhD Fellows. The Google PhD Fellowship is a scholarship program that supports graduate school students from around the world that have produced excellent achievements from promising computer science-related fields. The 75 selected fellows will receive ten thousand dollars of funding with the opportunity to discuss research and receive one-on-one feedback from experts in related fields at Google. Soo Ye Kim was recognized for her research outcomes in image and video quality enhancement. In particular, she was the first to suggest a deep learning-based method to restore super-resolution and HDR videos, and to handle super-resolutionization and interpolation at the same time. Her related research outcomes were presented at top international conferences in the fields of computer vision and AI, including CVPR, ICCV, and AAAI. She is also collaborating with Google and Adobe research teams through research internships to research various high-quality video conversion methods. Sanghyun Woo was recognized for his research in the fields of visual perception and deduction. He suggested an effective deep learning model design based on the human attention mechanism, and efficient learning methodologies utilizing self-learning and simulators, which received a lot of attention. His various research outcomes related to models and learning methodologies were presented at top international conferences in the fields of computer vision and AI, including CVPR, ECCV, and NeurlPS. In particular, his paper presented at ECCV in 2018, “Convolutional Block Attention Module (CBAM)”, is being used in various computer vision applications, and has exceeded 2700 citation counts on Google Scholar. Woo was also selected as a Microsoft Research Asia PhD Fellow in 2020. Hae Beom Lee’s research achievements include effectively overcoming various limitations of the existing meta-learning framework. Specifically, he proposed ways to deal with a realistic task distribution with imbalances, improved the practicality of meta-knowledge, and made meta-learning possible, even in large-scale task scenarios. These various studies have been accepted to numerous top-tier machine learning conferences such as NeurIPS, ICML, and ICLR. In particular, one of his papers was selected as an oral presentation at ICLR 2020 and another as a spotlight presentation at NeurIPS 2020. Due to the COVID-19 pandemic, the award ceremony was held via the online Google PhD Fellow Summit, and the list of recipients can be found on the Google website .
-
Deep Learning Framework to Enable Material Design ..
Researchers propose a deep neural network-based forward design space exploration using active transfer learning and data augmentation < Figure 1: Schematic of deep learning framework for material design space exploration. Schematic of gradual expansion of reliable prediction domain of DNN based on the addition of data generated from the hyper-heuristic genetic algorithm and active transfer learning. > A new study proposed a deep neural network-based forward design approach that enables an efficient search for superior materials far beyond the domain of the initial training set. This approach compensates for the weak predictive power of neural networks on an unseen domain through gradual updates of the neural network with active transfer learning and data augmentation methods. Professor Seungwha Ryu believes that this study will help address a variety of optimization problems that have an astronomical number of possible design configurations. For the grid composite optimization problem, the proposed framework was able to provide excellent designs close to the global optima, even with the addition of a very small dataset corresponding to less than 0.5% of the initial training data-set size. This study was reported in npj Computational Materials last month. “We wanted to mitigate the limitation of the neural network, weak predictive power beyond the training set domain for the material or structure design,” said Professor Ryu from the Department of Mechanical Engineering. Neural network-based generative models have been actively investigated as an inverse design method for finding novel materials in a vast design space. However, the applicability of conventional generative models is limited because they cannot access data outside the range of training sets. Advanced generative models that were devised to overcome this limitation also suffer from weak predictive power for the unseen domain. Professor Ryu’s team, in collaboration with researchers from Professor Grace Gu’s group at UC Berkeley, devised a design method that simultaneously expands the domain using the strong predictive power of a deep neural network and searches for the optimal design by repetitively performing three key steps. First, it searches for few candidates with improved properties located close to the training set via genetic algorithms, by mixing superior designs within the training set. Then, it checks to see if the candidates really have improved properties, and expands the training set by duplicating the validated designs via a data augmentation method. Finally, they can expand the reliable prediction domain by updating the neural network with the new superior designs via transfer learning. Because the expansion proceeds along relatively narrow but correct routes toward the optimal design (depicted in the schematic of Fig. 1), the framework enables an efficient search. As a data-hungry method, a deep neural network model tends to have reliable predictive power only within and near the domain of the training set. When the optimal configuration of materials and structures lies far beyond the initial training set, which frequently is the case, neural network-based design methods suffer from weak predictive power and become inefficient. Researchers expect that the framework will be applicable for a wide range of optimization problems in other science and engineering disciplines with astronomically large design space, because it provides an efficient way of gradually expanding the reliable prediction domain toward the target design while avoiding the risk of being stuck in local minima. Especially, being a less-data-hungry method, design problems in which data generation is time-consuming and expensive will benefit most from this new framework. The research team is currently applying the optimization framework for the design task of metamaterial structures, segmented thermoelectric generators, and optimal sensor distributions. “From these sets of on-going studies, we expect to better recognize the pros and cons, and the potential of the suggested algorithm. Ultimately, we want to devise more efficient machine learning-based design approaches,” explained Professor Ryu.This study was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research Project. -Publication Yongtae Kim, Youngsoo, Charles Yang, Kundo Park, Grace X. Gu, and Seunghwa Ryu, “Deep learning framework for material design space exploration using active transfer learning and data augmentation,” npj Computational Materials (https://doi.org/10.1038/s41524-021-00609-2) -Profile Professor Seunghwa Ryu Mechanics & Materials Modeling Lab Department of Mechanical Engineering KAIST

